
Once styles are defined, it’s far quicker to format objects by applying a style than by applying ad hoc formats for font, font size, field color, field alignment, padding, etc.
#Filemaker pro 11 change menu bar pro#
The "Requests" Menu) and clicking on the " Omit"Īppears by clicking on a field while pressing the ' ctrl' key, allowing theīutton at the top of the window to start the query.Like character and paragraph styles in your favorite word processor, object-formatting styles in FileMaker Pro 13 and FileMaker Pro 13 Advanced are double-barreled time-savers. Mode, the "AND" - "OR" - "NOT" operators may be implemented in this way: "requests" that are made for searching the database. The mode pop-up Menu bar at the bottom left of theįorm allowing to search es in specific fields.īook icon in the upper left corner represents different Pro-based database may be used basically in these "modes":ĭifferent modes can be done from the ' View' Menu or from the pop-up Menu bar atįor specific content in the database fields, using any different On the green circular button will retrieve the complementary Records: total number of Records in the table.įound: total number of the subset of Records Of the window, by using the cursor to the right of the small bookįollowing information is constantly displayed on the Status Is independent from the storage of the contained data.Ĭan be done by clicking on the ' Layout:'ĭatabase by clicking on the small book pages at the top left Multiplication of each mRNA length for itsĮxpression value in condition "B" the sum of thisįields from a table or its related fields from other tables. The imported expression value for each mRNA in theĬalculate the multiplication of each GC count for Imported expression value for each mRNA in the Referred to its length the mean and the standard deviationĪbsolute number of GC for each mRNA sequence

Ĭalculate the percentage of GC for each mRNA This section, TGCA fields are described. Gene symbol, NM_Accession, Sequence and length.
Separated by the tabulator keyboard button (TAB, ACII19): "NM_ACCESSION", "Seq_Line" and "bp" fields from the left squareĪlternatively, it is possible to manually create a data sequence file with four columns In the appearing window, in the "Group by" square checkīoth "SYMBOLUM" and "bp". Order for the "bp" field) obtaining the situationĮxport sequences for the longest isoforms for each geneįor sorted records choosing " Export Records."
#Filemaker pro 11 change menu bar software#
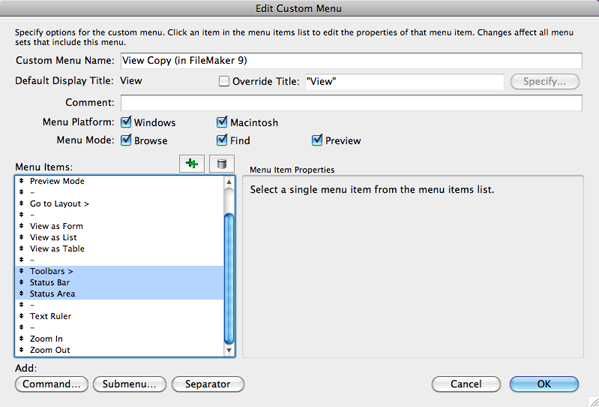
The software status toolbar, select the "SYMBOLUM" and "bp" fields from the left squareĪnd click on the "Move" button (select also Descending In order to export only the longest isoform for each gene, "5'_ORF_Extender.app" icon and go to the RefSeq_mRNA table Human Refseq database data already parsed through 5'_ORF_Extender are available at '_ORF_Extender/5'_ORF_Extender_2.0/human/Īfter the downloaded file decompression, open theĥ'_ORF_Extender software by double-clicking on the Updated data, please follow the 5'_ORF_Extender user guide. Order to obtain mRNA sequences, the use of theĪble to parse RefSeq database, is recommended.ĥ'_ORF_Extender is freely available for download at. Tabulator keyboard button (TAB, ACII19): Gene symbol,Įxpression values for "A" condition and Expression values Represented in the following figure and lastly click onĪlternatively, it is possible to manually create anĮxpression data file with three columns separated by the "Expression_A" and "Expression_B" fields from the left squareĪnd click on the "Move" button obtaining the situation In the appearing window, select the "Gene_name", Records choosing " Export Records." from the " File"Ĭhoose the type "Tab-Separate Text" and name the file as Represents DS AMKL and "B" represents MK) for retrieved

On "Omit" and then on "Perform Find" buttons:Įxport genes and the corresponding expression values ("A" Toolbar, type the "=" character in the "Ratio of In order to find genes with available expression valuesįor both conditions, click on the button of the software status To the "Values_A_B" software table by clicking on Software by double-clicking on the "TRAM.app" icon and go Maps from Acute Megakaryoblastic Leukemia (AMKL) inĬhildren with Down Syndrome (DS) and from normalĬhoose "Mac" or "Win" folder depending on your operatingĪfter the downloaded file decompression, open the TRAM (folders: 2014_Caracausi, 2014_Pelleri, 2015_Caracausi,Ģ016_Caracausi, for details please see the readme.htmlįile and the corresponding publications). Transcriptome maps already generated through TRAM for ĭesired transcriptome maps, please follow the TRAM user
TRAM is freely available for download at. To generate and analyse transcriptome maps, is Expression values, the use of the TRAM (Transcriptome


 0 kommentar(er)
0 kommentar(er)
